New paper from our laboratory
Sugihara, Y., Abe, Y., Takagi, H., Abe, A., Shimizu, M., Ito, K., Kanzaki, E., Oikawa, K., Kourelis, J., Langner, T., Win, J., Białas, A., Lüdke, D., Contreras, M. P., Chuma, I., Saitoh, H., Kobayashi, M., Zheng, S., Tosa, Y., Banfield, M., Kamoun, S., Terauchi, R., & Fujisaki, K. (2023). Disentangling the complex gene interaction networks between rice and the blast fungus identifies a new pathogen effector. PLOS Biology, 21(1), e3001945-. https://doi.org/10.1371/journal.pbio.3001945
Abstract
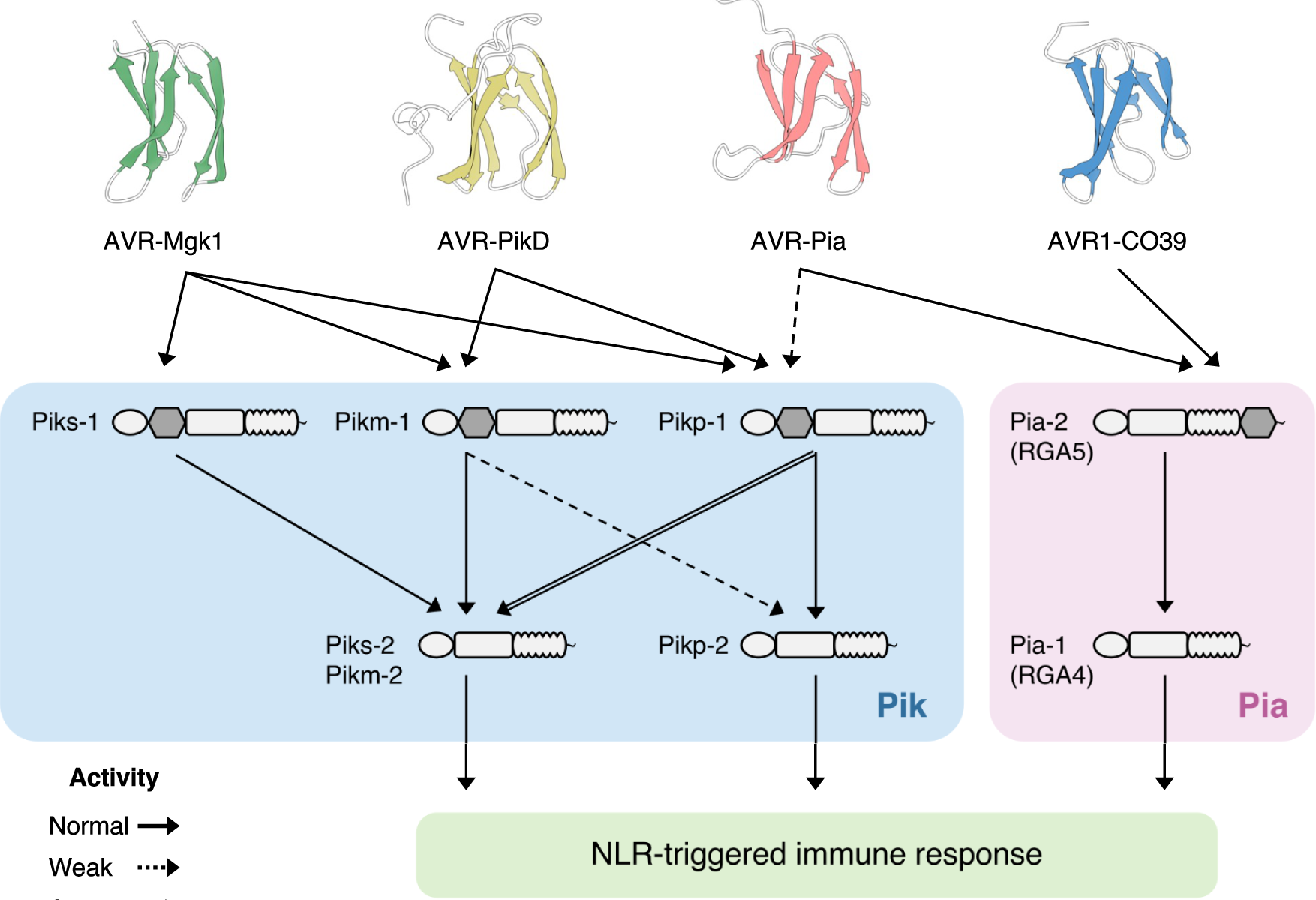
Studies focused solely on single organisms can fail to identify the networks underlying host–pathogen gene-for-gene interactions. Here, we integrate genetic analyses of rice (Oryza sativa, host) and rice blast fungus (Magnaporthe oryzae, pathogen) and uncover a new pathogen recognition specificity of the rice nucleotide-binding domain and leucine-rich repeat protein (NLR) immune receptor Pik, which mediates resistance to M. oryzae expressing the avirulence effector gene AVR-Pik. Rice Piks-1, encoded by an allele of Pik-1, recognizes a previously unidentified effector encoded by the M. oryzae avirulence gene AVR-Mgk1, which is found on a mini-chromosome. AVR-Mgk1 has no sequence similarity to known AVR-Pik effectors and is prone to deletion from the mini-chromosome mediated by repeated Inago2 retrotransposon sequences. AVR-Mgk1 is detected by Piks-1 and by other Pik-1 alleles known to recognize AVR-Pik effectors; recognition is mediated by AVR-Mgk1 binding to the integrated heavy metal-associated (HMA) domain of Piks-1 and other Pik-1 alleles. Our findings highlight how complex gene-for-gene interaction networks can be disentangled by applying forward genetics approaches simultaneously to the host and pathogen. We demonstrate dynamic coevolution between an NLR integrated domain and multiple families of effector proteins.
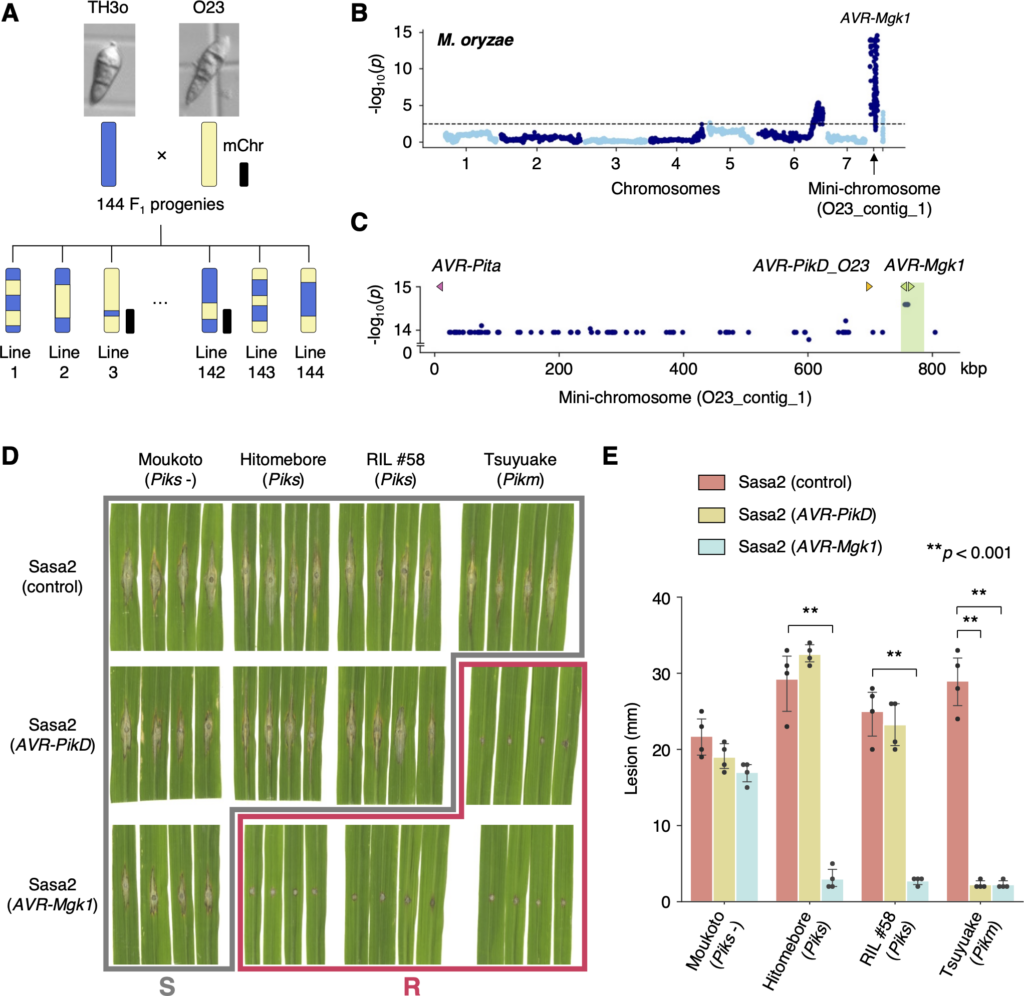
(A) Schematic representations of the F1 progeny generated after a cross between M. oryzae isolates TH3o and O23. We subjected all F1 progeny to whole-genome sequencing. O23 possesses a mini-chromosome. (B) Genetic association of the TH3o × O23 F1 progeny using infection lesion size on RIL #58 (Pish -, Piks +) rice plants as a trait. The vertical axis indicates -log10(p), where the p-value is how likely the marker shows association with a trait due to random chance. The dashed line shows the p-value corresponding to a false discovery rate of 0.05. The association analysis based on the O23 reference genome identified AVR-Mgk1, encoded on the mini-chromosome sequence O23_contig_1, as an AVR gene. O23_contig_1 was not present in the TH3o genome and was unique to the O23 genome. We used 7,867 SNP markers for chromosomes 1–7 and 265 presence/absence markers for the other contigs. (C) p-values for O23_contig_1 with annotated AVRs. We also detected AVR-Pita and AVR-PikD in O23_contig_1. AVR-PikD in O23_contig_1 contains a frameshift mutation, so we named this variant AVR-PikD_O23. The region encoding 2 AVR-Mgk1 genes and showing lower p-values is highlighted in green. Nucleotide sequences of the 2 AVR-Mgk1 genes, arranged in a head-to-head orientation, are identical. (D) Results of punch inoculation assays using M. oryzae isolate Sasa2 transformed with AVR-PikD or AVR-Mgk1. Wild-type Sasa2 infected all the cultivars tested in this study. The Sasa2 transformant expressing AVR-PikD infected RIL #58 (Piks), but that expressing AVR-Mgk1 did not infect RIL #58 (Piks) or Tsuyuake (Pikm) rice plants. (E) Quantification of the lesion size in (D). Asterisks indicate statistically significant differences (p < 0.001, two-sided Welch’s t test). The data underlying Fig 4B and 4C and 4E can be found in S1 Data. AVR, avirulence; RIL, recombinant inbred line; SNP, single nucleotide polymorphism. https://doi.org/10.1371/journal.pbio.3001945.g004