REVIEW ARTICLE
Kourelis, J., & Adachi, H. (2022). Activation and Regulation of NLR Immune Receptor Networks. Plant and Cell Physiology, pcac116. https://doi.org/10.1093/pcp/pcac116
Abstract
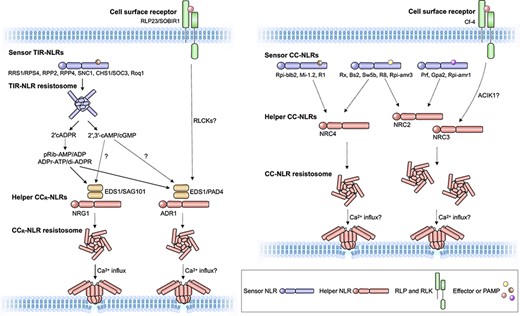
Plants have many types of immune receptors that recognize diverse pathogen molecules and activate the innate immune system. The intracellular immune receptor family of nucleotide-binding domain leucine-rich repeat–containing proteins (NLRs) perceive translocated pathogen effector proteins and execute a robust immune response, including programmed cell death. Many plant NLRs have functionally specialized to sense pathogen effectors (sensor NLRs) or to execute immune signalling (helper NLRs). Sub-functionalized NLRs form a network-type receptor system known as the NLR network. In this review, we highlight the concept of NLR networks, discussing how they are formed, activated, and regulated. Two main types of NLR networks have been described in plants: the ADR1/NRG1 network and the NRC network. In both networks, multiple helper NLRs function as signalling hubs for sensor NLRs and cell surface–localized immune receptors. Additionally, the networks are regulated at the transcriptional and posttranscriptional levels, as well as being modulated by other host proteins to ensure proper network activation and prevent autoimmunity. Plant pathogens in turn have converged on suppressing NLR networks, thereby facilitating infection and disease. Understanding the NLR immune system at the network level could inform future breeding programs by highlighting the appropriate genetic combinations of immunoreceptors to use while avoiding deleterious autoimmunity and suppression by pathogens.