New paper from our laboratory
Kovi, B., Sakai, T., Abe, A., Kanzaki, E., Terauchi, R., & Shimizu, M. (2023). Isolation of Pikps, an allele of Pik, from the aus rice cultivar Shoni. Genes & Genetic Systems, advpub. https://doi.org/10.1266/ggs.22-00002
Abstract
Blast disease caused by the filamentous fungus Pyricularia oryzae (syn. Magnaporthe oryzae) is one of the most destructive diseases of rice (Oryza sativa L.) around the globe. An aus cultivar, Shoni, showed resistance against at least four Japanese P. oryzae isolates. To understand Shoni’s resistance against the P. oryzae isolate Naga69-150, genetic analysis was carried out using recombinant inbred lines developed by a cross between Shoni and the japonica cultivar Hitomebore, which is susceptible to Naga69-150. The result indicated that the resistance was controlled by a single locus, which was named Pi-Shoni. A QTL analysis identified Pi-Shoni as being located in the telomeric region of chromosome 11. A candidate gene approach in the region indicated that Pi-Shoni corresponds to the previously cloned Pik locus, and we named this allele Pikps. Loss of gene function mediated by RNA interference demonstrated that a head-to-head-orientated pair of NBS-LRR receptor genes (Pikps-1 and Pikps-2) are required for the Pikps-mediated resistance. Amino acid sequence comparison showed that Pikps-1 is 99% identical to Pikp-1, while Pikps-2 is identical to Pikp-2. Pikps-1 had one amino acid substitution (Pro351Ser) in the NBS domain as compared to Pikp-1. The recognition specificity of Pikps against known AVR-Pik alleles is identical to that of Pikp.
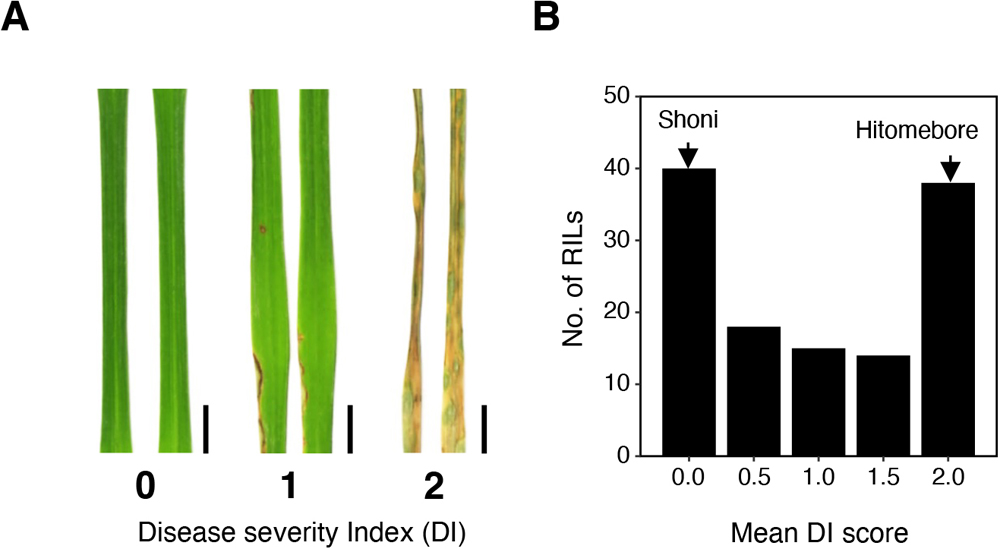
(A) The disease severity index (DI) was employed in evaluating phenotypes of RILs after spray inoculation of the fungus. DI = 0, no symptoms; DI = 1, 0 to 20% infected leaf area; DI = 2, over 20% infected leaf area. Representative leaves for each category are shown. Scale bars, 0.5 cm. (B) Frequency distribution of the DI for 125 RILs derived from a cross between Hitomebore and Shoni. Arrows indicate approximate value obtained for the parental (Hitomebore and Shoni) lines. The DI score of each RIL is represented by the average value of two technical replications.
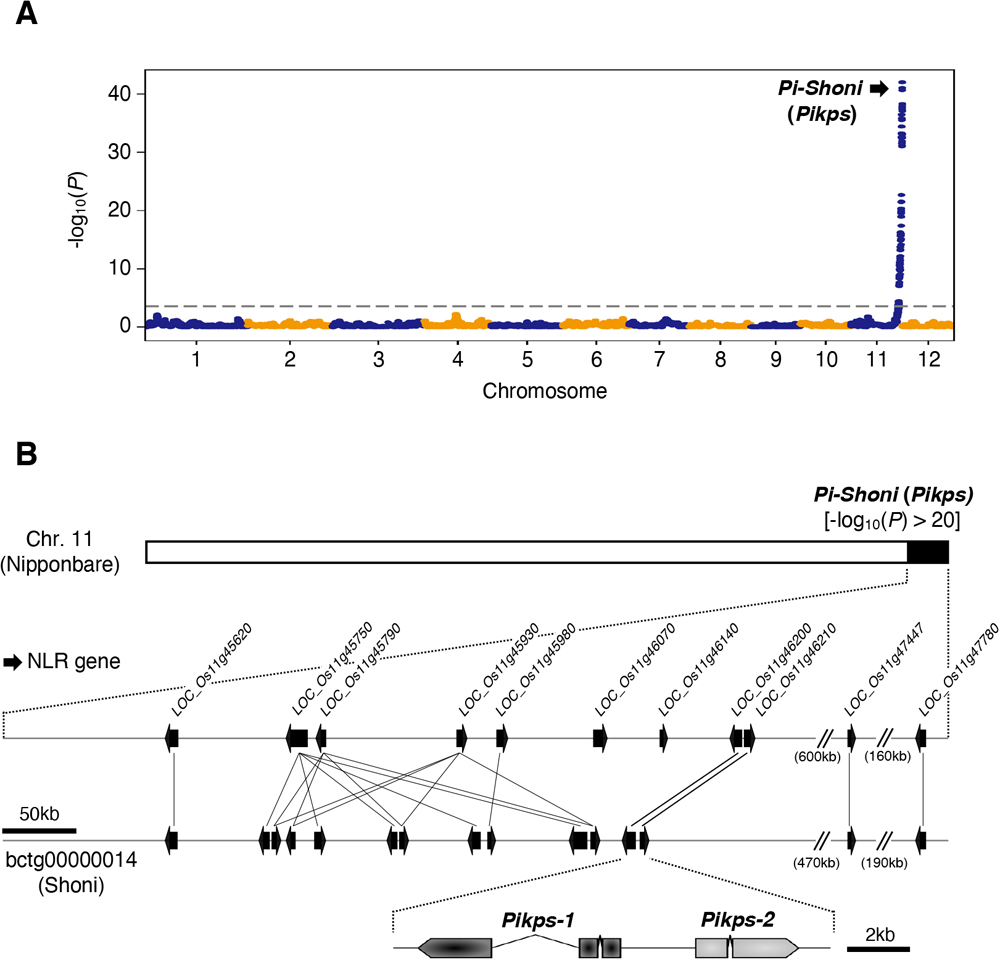
(A) QTL analysis of DI scores obtained from the 125 RILs. The dashed line indicates the significance threshold (-log10(P) > 3.36). (B) Comparative genomic mapping of the 11 NLR genes in Nipponbare within the scaffold bctg00000014 of the Shoni genome assembly. The black arrows indicate NLR genes. Pikps-1 and Pikps-2 correspond to LOC_Os11g46200 and LOC_Os11g46210, respectively